Problem 34: Genes can be moved between species.
Use green fluorescent protein to tag expression of genes.
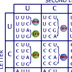
HI! Green fluorescent protein (GFP) is a protein that causes the Aequorea victoria jellyfish to glow. The protein is coded for by a single gene. The GFP gene can be inserted downstream of the promoter of a gene in another organism. RNA polymerase binds to promoter regions to initiate transcription. If the GFP gene is inserted correctly, it can be expressed in organisms other than jellyfish. The GFP gene can be used as a visual tag for the expression of other genes. You want to look at the expression of the -galactosidase gene, which produces -galactosidase, an enzyme that digests lactose. To do this, you're going to fuse -gal to the GFP gene. You are going to use a plasmid X for your experiment. It is 8 kb and already has -gal on it, as well as an ampicillin resistance gene. Where on this plasmid would be the best place to insert the GFP gene? A (No, this is far from the end of the -gal gene.) B (No, inserting the GFP gene here will destroy the ampicillin resistance gene.) C (No, this is in front (upstream) of the -gal promoter.) D (That is correct) Point D is between the promoter and the gene. DNA inserted here would be transcribed and translated as a fusion protein with the -galactosidase enzyme. Transcription begins at the -gal promoter and continues through the end of the -gal gene. If the GFP gene is inserted correctly, there should be no stop codons and a GFP- -gal fusion protein will be made from the mRNA. Remember the genetic code is read as triplet codons. Assuming that the inserted piece of DNA starts at the first position of the triplet codon, then in this stretch of plasmid DNA, where is the best place to insert DNA? 1 (That is correct) 2 (No, this will shift the reading frame for later amino acids.) 3 (No, this will shift the reading frame for later amino acids.) If DNA is inserted at the other positions, the reading frame is shifted and functional protein may not be made. You isolate and purify the GFP gene as a small piece of DNA around 1 kb. It has EcoRI "sticky" ends. You then mix the GFP piece with pX plasmids also cut with EcoRI. You add DNA ligase to ligate the pieces together. You then use the mix to transform bacteria ... ... and you grow the bacteria on media containing ampicillin to find those that accepted the plasmid and have been transformed. Under UV light, some bacteria glow and some do not. You isolate the plasmids and then cut them with EcoRI. You check the sizes of the plasmids by running them on a gel. Remember the GFP gene is about 1 kb and the pX plasmid is 8 kb. Why don't the plasmids in lane four glow? The GFP gene was inserted backwards. (That is correct) The plasmids recircularized. (No, the plasmid sizes add up to the size of pUC + GFP.) The promoter region was destroyed. (No, the promoter is fine.) None of the above. (No, there is a correct answer.) The size of the pieces on the gel indicates that both pX and GFP are present. Since GFP is not working, most likely the gene was inserted backwards. When you use a single enzyme to cut both a gene and a plasmid, three things can happen. The plasmid can recircularize. The plasmid can accept the new gene. Or the gene can be inserted backwards. The sticky ends left by enzymes are palindromes: they are the same forwards or backwards. Plasmids made for recombinant DNA technology have a region containing many enzyme restriction sites called a polylinker region. This allows many different cutting options to match a gene the scientist wants to insert. If you wanted to avoid the problem of inserting a gene backwards, which would be the best restriction enzymes to use? EcoRI (No, using a single enzyme the GFP gene can be inserted backwards or forwards.) PvuI and SalI (That is correct.) BamHI and HindIII (No, using these enzymes the GFP gene will be inserted backwards.) PvuI and XbaI. (No, this will generate two XbaI sites around the GFP.) Both the pX plasmid and the DNA with the GFP gene should be cut with the enzymes PvuI and SalI. The complementary ends made by the enzymes are such that the GFP gene can only be inserted in the forward orientation. CONGRATULATIONS!!! YOU'RE SO SMART!
gfp gene, ampicillin resistance, rna polymerase, stop codons, promoter regions, fusion protein, aequorea victoria, green fluorescent protein, resistance gene, plasmid dna, amino acids, ampicillin, first position, genetic code, jellyfish, triplet, lactose, genes, mrna
- ID: 16723
- Source: DNALC.DNAFTB
Related Content
16898. New York Stories: Martin Chalfie and Green Fluorescent Protein (GFP)
New York high school students interview Nobel Laureate, Dr. Martin Chalfie of Columbia University, then perform the experiment with green fluorescent protein (GFP) that he pioneered.
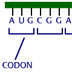
15545. Tanscription/translation - Start and stop codons
The diagram represents a single strand of DNA containing a gene, in purple. Remember this gene is "read" in the 5' to 3' direction to produce an mRNA.
16513. Problem 22: DNA words are three letters long.
Decode a protein.
16834. Animation 40: Living things share common genes.
Mike Wigler shows how all organisms share similar genes, called homologs.
16494. Animation 22: DNA words are three letters long.
Several researchers crack the genetic code.
16492. Problem 21: RNA is an intermediary between DNA and protein.
What happens in protein synthesis?
16705. Animation 34: Genes can be moved between species.
Stanley Cohen and Herbert Boyer transform bacteria with a recombinant plasmid, and Doug Hanahan studies induced transformation.
16514. Concept 23: A gene is a discrete sequence of DNA nucleotides.
Gene analysis take a giant leap using DNA sequencing.
16569. Problem 25: Some viruses store genetic information in RNA.
Explore the reverse transcriptase mechanism.