
Educator Training
We offer up-to-date teacher training through biology workshops and professional development for teachers in genetics and biotechnology. With federal and private foundation funding, we offer these free workshops to middle school, high school, and college educators, especially those in the areas of genetics, biology, genomics, and bioinformatics.
The Cold Spring Harbor Laboratory DNA Learning Center is an approved Sponsor of Continuing Teacher and Leader Education (CTLE).
Nanopore Sequencing for Student Research
Saturday, April 18, 2026
9:30 a.m.–3:00 p.m.
DNA Learning Center NYC at City Tech
Brooklyn, NY
Saturday, April 25, 2026
9:30 a.m.–3:00 p.m.
Dolan DNA Learning Center
Cold Spring Harbor, NY
Saturday, May 16, 2026
9:30 a.m.–3:00 p.m.
Regeneron DNA Learning Center
Sleepy Hollow, NY
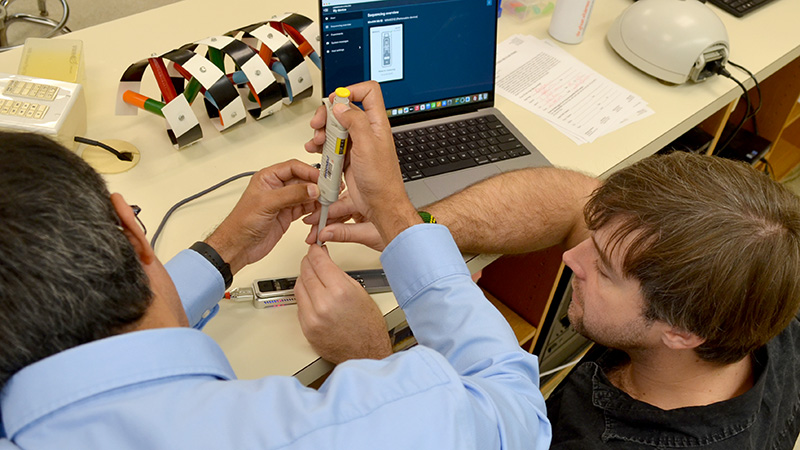
Join us this spring to see how nanopore sequencing—a portable, cost-effective alternative to traditional commercial services—can be integrated into your classroom. This workshop demonstrates how to use nanopore technology for real-world applications, including generating DNA barcodes and performing microbiome or metabarcoding analyses.
The DNA Learning Center and Oxford Nanopore have collaborated to adapt these professional workflows for education, streamlining the protocol to ensure the technology is accessible for student use. Additionally, we will showcase the redeveloped DNA Subway 2.0, which now supports full mobile use and a dedicated line for metabarcoding analysis, enabling users to “analyze anywhere”.
Finally, attendees will learn how the DNALC’s Nanopore Research Accelerator partnership can offer ongoing support for implementing these projects in their own schools.
- In-person free workshop
- Saturday, April 18, 2026
DNA Learning Center NYC at City Tech
62 Tillary Street, Brooklyn, NY 11201 - Saturday, April 25, 2026
Dolan DNA Learning Center
334 Main St, Cold Spring Harbor, NY 11724 - Saturday, May 16, 2026
Regeneron DNALC
1 Rockwood Road Sleepy Hollow, NY 10591 - Directions
Past Event
DNA Subway 2.0 Educator Webinar
With the legacy platform retiring in June 2025, this workshop guides educators through the essential transition to DNA Subway 2.0. We will conduct a live walkthrough of the new mobile-first interface, demonstrate the expanded Blue Line capabilities for Oxford Nanopore sequencing, and cover the practical steps for migrating your curriculum. Join us to ensure your lab materials are updated and ready for the next academic year.
Join Jason Williams for a live, virtual demonstration of DNA Subway 2.0, with a walk-through of the new interface, enhanced Blue Line analysis tools, and more. This is your chance to ask questions, troubleshoot, and receive personalized guidance on how to integrate DNA Subway 2.0 into your teaching.
Virtual free workshop:
- Wednesday, February 18, 2026
4:00 p.m.–5:00 p.m. Eastern Time OR - Tuesday, February 24, 2026
7:00 p.m.–8:00 p.m. Eastern Time - A Zoom link will be provided to participants after registration
Webinar recording:
Sites of Major DNALC Faculty Workshops, 1985-2014
This map shows the locations of the DNALC's faculty workshops taught over more than thirty years.
Open the map key map key to show/hide the years in groups of three. Click the check boxes to show or hide the years. Click the dots for information on host institution, year, and instructional level of participating faculty. Map can be opened full screen in a separate browser window by clicking the full screen icon at the upper-right.


