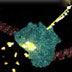
The cycles of the polymerase chain reaction (PCR), 3D animation
Until the mid-1980s, the only way to make many copies of DNA was to insert the DNA pieces into bacteria and select the desired one from many different colonies growing on a plate. In 1985, Kary Mullis invented a precise and radical new method of selecting and amplifying a section of DNA ? the polymerase chain reaction (PCR). (DNAi Location: Manipulation > Techniques > Amplifying > PCR animation)
Polymerase chain reaction, or PCR, uses repeated cycles of heating and cooling to make many copies of a specific region of DNA. First, the temperature is raised to near boiling, causing the double-stranded DNA to separate, or denature, into single strands. When the temperature is decreased, short DNA sequences known as primers bind, or anneal, to complementary matches on the target DNA sequence. The primers bracket the target sequence to be copied. At a slightly higher temperature, the enzyme Taq polymerase, shown here in blue, binds to the primed sequences and adds nucleotides to extend the second strand. This completes the first cycle. In subsequent cycles, the process of denaturing, annealing and extending are repeated to make additional DNA copies. After three cycles, the target sequence defined by the primers begins to accumulate. After 30 cycles, as many as a billion copies of the target sequence are produced from a single starting molecule.
target dna sequence,polymerase chain reaction,polymerase chain reaction pcr, 3d animationkary mullis,dna amplification,dna strands,target sequence,dna pieces,dna sequences,dna copies,double stranded dna,manipulation techniques,mid 1980s,dnai,chain reaction,annealing,primers,molecule,bacteria,bind
- ID: 15475
- Source: DNALC.DNAi
- Download: MPEG 4 Video
Related Content
15625. Polymerase chain reaction (PCR)
DNA polymerase (blue) makes many copies of DNA (red) in a cycle of the polymerase chain reaction (PCR).
17044. Polymerase chain reaction (PCR)
Polymerase chain reaction (PCR) enables researchers to produce millions of copies of a specific DNA sequence in approximately two hours. This automated process bypasses the need to use bacteria for amplifying DNA.
15479. Sanger method of DNA sequencing, 3D animation with narration
The DNA sequencing method developed by Fred Sanger forms the basis of automated "cycle" sequencing reactions today.
15138. Naming PCR
Kary Mullis explains how the polymerase chain reaction (PCR) was named.
15140. Making many DNA copies, Kary Mullis
Kary Mullis talks about his discovery of the polymerase chain reaction (PCR), a process that allows chemists to produce many copies of a specific fragment of DNA.
15624. Kary Mullis
Image of Kary Mullis. In 1985, Kary Mullis invented the polymerase chain reaction (PCR), a method of amplifying or producing many copies of a specific piece of DNA. The revelation came to this eccentric character on a drive in northern California.
15139. Finding DNA to copy, Kary Mullis
Kary Mullis speaks about the process of find a specific fragment of DNA amongst many pieces in a complex mixture.
16515. Animation 23: A gene is a discrete sequence of DNA nucleotides.
Fred Sanger outlines DNA sequencing.
16065. Kary Mullis
KARY MULLIS (1944- )
15480. Transcription: DNA to RNA, 3D animation with sound effects only
Transcription factors bind to DNA, RNA polymerase begins transcribing messanger RNA (mRNA) molecule from DNA.