Some DNA does not encode protein.
Explore Britten's DNA reassociation rates for different organisms.
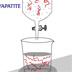
HI! Britten’s Cot curve analysis compares DNA complexity. A useful number in this type of analysis is Cot1/2. This is the value when 50% of the DNA has reassociated. For example, the Cot1/2 of E. coli is 9. What is the approximate Cot1/2 of bacteriophage T4 DNA? 9 (No, this is the Cot1/2 of E. coli.) 1 (No, this is not correct.) 10 (No, this is close to the Cot1/2 of E. coli.) 10 -1 (That is correct) The Cot1/2 of T4 DNA is 3 X 10-1. The Cot1/2 value is derived from the log of the DNA concentration X time. A greater Cot1/2 value means a slower reaction. It takes longer for E. coli DNA to completely reassociate than bacteriophage T4. The Cot1/2 value is also related to the amount of DNA present in the genome — the concentration of complementary sequences. A larger genome has more DNA, but by comparing Cot1/2 values with E. coli, the "complexity" of a genome can be calculated. The complexity is the total length of different sequences present in a genome and is usually expressed in base pairs. The E. coli genome has only unique sequences; the total length of different sequences is 4.2 X 106bp. What is the complexity of bacteriophage T4 DNA? 4.2 X 10 6 bp (No, this is the size of the E. coli genome.) 4.2 X 10 5bp (No, this is not correct.) 1.4 X 10 6 bp (No, this is not correct.) 1.4 X 10 5 bp (That is correct) Bacteriophage T4 has a Cot1/2 of 3 X 10 -1, and the Cot1/2 of E. coli is 9. The complexity of the bacteriophage T4 genome is 1.4 X 10 5bp. Now, let’s look at eukaryotic DNA. Since there are three components to this reassociation curve, each component has its own Cot1/2value. What are the Cot1/2values of the fast, medium and slow components? 0.004, 3, 1000 (That is correct) 10, 10, 10 (No, the Cot1/2 values aren’t all the same.) 0.004, 10, 1000 (No, one of the values is incorrect.) 1, 10, 1000 (No, two of the values are incorrect.) The Cot1/2values of the fast, medium and slow components of this curve are 0.004, 3 and 1000. The fast component of this curve represents 25% of the total DNA, the medium component is 30% and the slow component is 45%. If we could isolate these fractions and run them as separate reassociation reactions, then the Cot1/2values would be: 0.004, 3, 1000 (No, these are the original Cot values. 0.25, 0.3, 0.45 (No, these are the % DNA in each component.) 1 X 10 -3, 0.9, 450 (That is correct) If we treat each of the components as separate reactions, then the Cot1/2values for the fast, medium and slow curves would be 1 X 10 -3, 0.9, 450. What is the complexity of the slow fraction of this eukaryotic DNA? 1.47 X 10 8 bp. (No, that is not correct.) 4.7 X 10 8 bp. (That is correct) 5.95 X 10 8 bp. (No, that is not correct.) 6.62 X 10 8 bp. (No, that is not correct.) Since you are comparing the Cot1/2value of 45% of the eukaryotic DNA, the E. coli Cot1/2 value needs to be factored for 45%. This means that:450 / (9 X 0.45) = complexity of the slow fraction / 4.2 X 10 6= 4.7 X 10 8. The complexity of the slow fraction is the total length of unique sequence in this genome. Since the slow fraction is only 45% of the DNA, what is the estimated size of the whole genome? 4.7 x 10 8 bp. (No, this is not correct) 1.47 x 10 8 bp. (No, this is not correct) 1.0 x 10 9 bp. (That is correct) If 4.7 X 10 8 bp represents 45% of the genome, then 1.0 X 10 9 bp represents 100% of the genome. CONGRATULATIONS!!! YOU'RE SO SMART!
dna reassociation, dna complexity, dna concentration, complementary sequences, britten, cot curve analysis
- ID: 16666
- Source: DNALC.DNAFTB
Related Content
16657. Some DNA does not encode protein.
DNAFTB Animation 31: Roy Britten presents his work with David Kohne on repetitive DNA and its evolutionary origins.
16661. Video 31: Roy J. Britten, clip 1
Using mouse satellite DNA to study reassociation kinetics.
16027. Roy Britten, 1960
Some DNA does not encode protein.
15917. Cutting and pasting DNA
The discovery of enzymes that could cut and paste DNA made genetic engineering possible.
15573. DNA sequencer
An image of a DNA sequencer.
16663. Video 31: Roy J. Britten, clip 3
Composition of repeated DNA in the genome.
16664. Video 31: Roy J. Britten, clip 4
Are repeated DNA species specific?
15582. DNA fingerprint
DNA profiling, DNA fingerprinting, gel analysis
16665. Biography 31: Roy John Britten (1919 - )
Roy Britten did seminal research on repetitive DNA and its evolutionary origins.