Fulfilling the Promise of Nanopore Sequencing in Education
The Banbury Center
Cold Spring Harbor Laboratory
September 21–24, 2024
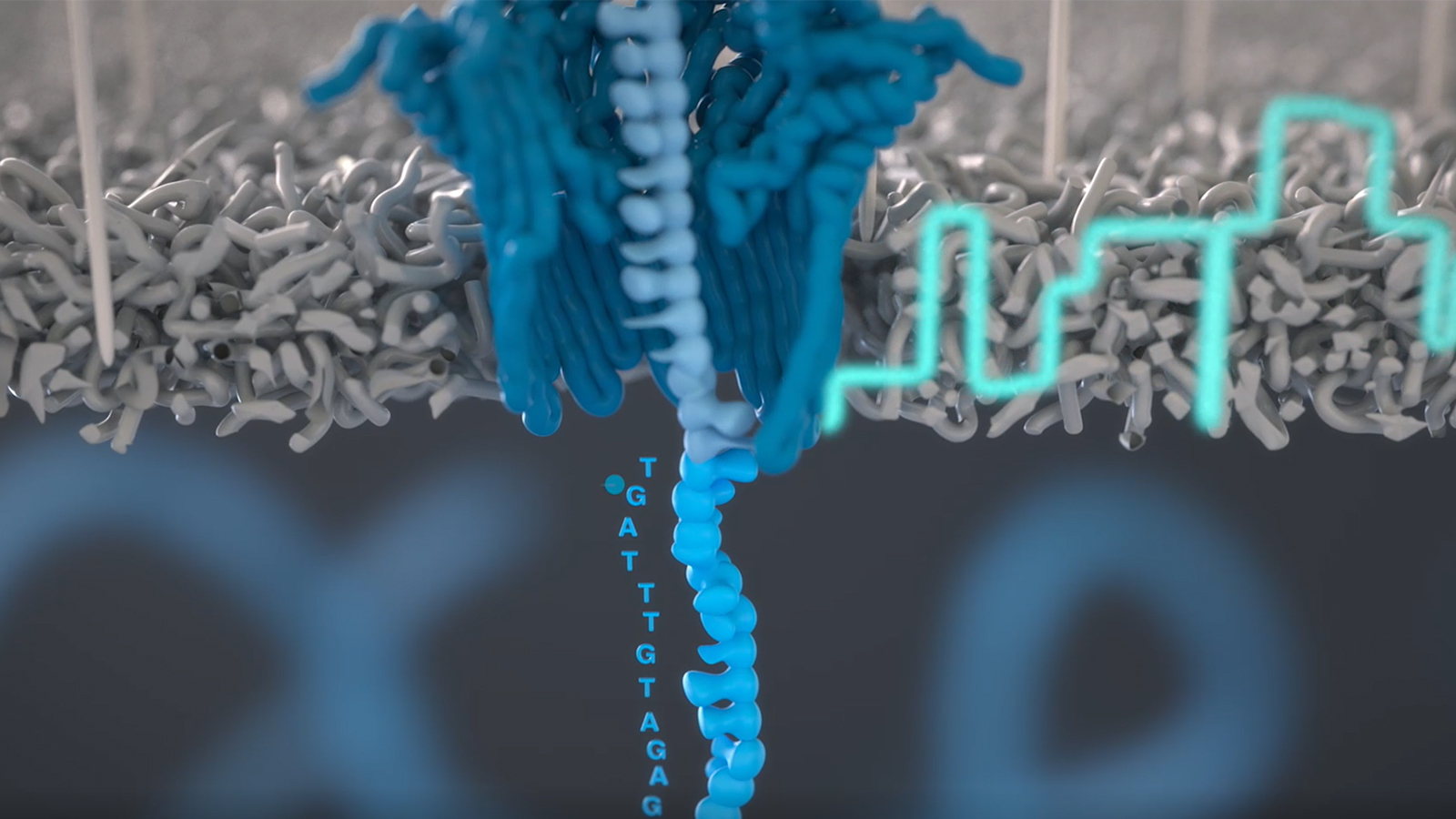

Nanopore sequencing technology offers inexpensive, real-time analysis of individual DNA molecules—potentially making DNA sequencing available anytime, anyplace, to anyone. This technology holds particular promise in bioscience education, where we envision a miniature Nanopore sequencer in every teaching lab, with students at all levels generating and exploring meaningful data.
For these reasons, we held a small meeting, “Fulfilling the Promise of Nanopore Sequencing in Education,” on September 21–24 at the Banbury Center of Cold Spring Harbor Laboratory. Against this backdrop, 25 scientists and educators met at Cold Spring Harbor Laboratory’s Banbury Center in September to discuss how to fulfill this promise of nanopore sequencing in education. During the meeting, we learned of large existing audiences of students who could readily use nanopore sequencing for projects on DNA barcoding (popularized by the DNALC ), bacteriophage analysis (popularized by the Howard Hughes Medical Institute), and identification of antibiotic-producing bacteria (popularized by the University of Wisconsin). We also heard great examples of nanopore class projects – from assessing microbial diversity in local waterways to cataloging plants visited by bees, to sequencing daffodil chloroplast genomes.
Attendees joined break out groups to discuss the promises and challenges of Nanopore sequencing, as well as proposals for broad implementation. Key themes included sustained training, resource accessibility, and community collaboration. Continuous mentorship, including virtual teaching assistants and "train-the-trainer" programs were seen as vital for long-term success. Participants highlighted the importance of creating user-friendly kits and clear protocols to make genomic research accessible across educational levels. Partnerships among educational institutions, industry, and community organizations are crucial for sharing resources and fostering a genomics-literate workforce. The DNALC, which signed an MOU with Oxford Nanopore last year, will continue working with the organizations represented at the meeting as we lay groundwork for popularizing this technology into the classroom.
This meeting is supported by grants from the National Science Foundation: Improving Undergraduate STEM Education (#1821657), and Advanced Technological Education (#1901984). Additional travel support is provided by Oxford Nanopore Technologies.
Organized by:
Jose L. Agosto-Rivera
STEM graduate training is typically centered around disciplinary research. Although teaching experiences are usually part of graduate training, they are viewed as peripheral experiences, even though a large proportion of Ph.D. students will end up in primarily undergraduate institutions (PUIs) or community colleges that mainly focus on teaching. This is partially due to the need for more integration between teaching and research and the fact that the primary determinant of graduation is the generation of new knowledge through research. In the past few years, all major educational agencies and authorities have encouraged the development of Course- Based Undergraduate Research Experiences (CUREs), which nicely integrate teaching and research. Nanopore sequencing technology represents an ideal tool for CUREs not only because of its cost-effectiveness but also because its versatility allows the integration of projects that range from ecology and environmental sciences to hardcore molecular biology and biomedical questions. Here, we present a demonstration project where we used nanopore amplicon sequencing in the undergraduate genetics laboratory to characterize bacterial species in beeswax that may serve as probiotics to facilitate the development of circadian circuits in the honey bee brain.
Dr. Jose L. Agosto-Rivera is an Associate Professor at the University of Puerto Rico, Rio Piedras Campus, and is currently the interim director of the Department of Biology. He has extensive scientific research experience in neurobiology and circadian rhythms. He has also been the coordinator of the Undergraduate Laboratory of Genetics for the last 8 years, where he has been integrating the research of the graduate students serving as teaching assistants into the curriculum of the laboratory. For the past 2 years, he has been collaborating with Prof. Jason Williams from Cold Spring Harbor in several research grants that involve integrating nanopore technology into undergraduate teaching laboratories.
Stokes S. Baker
Nanopore sequencing technologies can foster educational innovations such as course-based undergraduate research experience (CURE) pedagogy. Since 2019, nanopore sequencing has been used in Applied Metagenomics, a CURE that is part of the University of Detroit Mercy's (UDM) Building Infrastructure Leading to Diversity (http://rebuildetroit.org/) curriculum. Students enrolled in this course have contributed to science by publishing their data in a peer-reviewed manuscript (https://doi.org/10.3389/fmicb.2021.579325) and by uploading sequences to databases. An assessment study showed that students felt they were engaged as scientists while taking the course. Because of its flexibility, nanopore sequencing can be used to support CURE courses that investigate community-focused issues, such as urban park ecology. For the past two years, UDM undergraduates have conducted metabarcoding studies of urban beehives. The long-read capabilities were used to sequence 2.46 kbp amplicons containing both 18S and ITS rRNA sequences to analyze pollen and fungal profiles in honey. This fall, undergraduates will be conducting a metabarcoding study of Megachile rotundata, a solitary bee that is nearly stingless. M. rotundata colonies can be established inexpensively and in settings not appropriate for honeybees. Thus, community-based studies involving barcoding and metabarcoding can be conducted by diverse undergraduate institutions, including minority-serving institutions like UDM.
Stokes S. Baker teaches a course-based research experience (CURE) course entitled Applied Metagenomics and is developing CURE curricula involving nanopore-based investigations of pollinator biology. He also teaches courses in ecology and biostatistics. He has taught undergraduates at the University of Detroit Mercy for over 30 years. His disciplinary research uses molecular tools to investigate ecological questions like plantmicrobe interactions with duckweed. During the winter of 2024, he was a Fulbright Scholar at the University of Belize where he studied sea cucumber microbiomes.
Karen Barnard-Kubow
Projects centered on DNA sequencing are increasingly incorporated into undergraduate classes. While most steps of these projects are done in the classroom, sequencing itself is consistently outsourced, resulting in a “black box” of sequencing. Oxford Nanopore’s MinION sequencer offers a way to combat this “black box” by enabling sequencing to be carried out in the classroom due to its relatively low cost and small size. While it is assumed that sequencing in the classroom will have a positive impact on students, this has rarely been tested. We developed a short CURE-like module that exposes students in JMU’s core genetics lab to MinION sequencing. This module was implemented in 10 lab sections of 24 student each in fall 2023 and spring 2024. To test the impact of students interacting with the sequencer, the MinION sequencer was only present in half the lab sections. Post-module surveys were given to students to test for a difference in impact due to sequencer presence. Overall, students reported that the module had a positive impact. While there was an effect of sequencer presence, this was more pronounced in some groups of students than others. We also gained valuable experience in implementing MinION sequencing in the classroom.
Karen Barnard-Kubow. I am an evolutionary biologist interested in how the diversity of life we see around us is generated and maintained. My research lab currently addresses two main questions. First, what are the processes that drive genetic divergence between populations, leading to the formation of new species, where we focus on cytonuclear coevolution and incompatibility in the herb Campanula americana. Second, what are the environmental factors that structure zooplankton communities in riverine rock pools, generating and maintaining genetic and functional biodiversity. For this work we focus on characterizing zooplankton communities across time and space in the James River rock pools at Belle Isle in Richmond, VA. I am also the director of JMU’s Center for Genome and Metagenome Studies, whose mission is to support innovative research and training in the methods and principles of genomics, metagenomics, and bioinformatics to provide an exemplary learning experience for undergraduate and graduate students. As part of this role, I develop and implement genomic/bioinformatic modules to incorporate into our lab courses and train faculty and students on the use of various genomic and bioinformatic techniques and technologies.
Ellen Carpenter
The NSF vision is for a nation that leads the world in science and engineering research and innovation, to the benefit of all, without barriers to participation. This vision is achieved by NSF’s investments in research and education to support the NSF core values of scientific leadership, diversity and inclusion, integrity and excellence, public service, and innovation and collaboration. Engaging students in cutting-edge research in genomics and bioinformatics has the promise of both generating new scientific knowledge and training the scientific and technical workforce of the future. Support for science education across the spectrum “from K to gray” is available through multiple avenues at NSF. Biology education and biology education research are supported through the Directorates for Biology and STEM Education in particular, while cross-cutting fields such as data science, bioinformatics, behavioral science, and workforce development are supported by multiple directorates and divisions. Projects at single institutions as well as broad national networks all have a role to help enact NSF’s vision of a nation that leads the world in research, education, and innovation.
Dr. Ellen Carpenter is the lead program officer for the Improving Undergraduate STEM Education (IUSE) program in the Division of Undergraduate Education at the National Science Foundation. The IUSE program supports over 700 active awards focused on identifying what works, for whom, and in what contexts to improve STEM education for all undergraduate students. In addition to her work on the IUSE program, Dr. Carpenter has helped to develop and manage the STEM Education Postdoctoral Fellowship program, the Research Coordination Networks in Undergraduate Biology Education program, the Hispanic-Serving Institutions program, the Integrative Strategies in Neural and Cognitive Sciences program, and the Understanding the Rules of Life NSF Big Idea. Prior to joining the NSF, Dr. Carpenter was Professor of Psychiatry at the David Geffen School of Medicine and chair of the Neuroscience Undergraduate Interdepartmental Program at UCLA. She received her undergraduate degree in Biology from Dartmouth College, her doctoral degree in Neurobiology from the University of Chicago, and completed postdoctoral training in Human Genetics at the University of Utah.
Yvonne Chan
The ‘Āina-Informatics Network is a genomics education outreach program based at ‘Iolani School (Honolulu, HI) designed to bring authentic research experiences into Hawai’i grade 7-12 classrooms. Since 2018, the program has sequenced dozens of novel bacterial genomes, hundreds of SARS-CoV-2 genomes, environmental DNA, microbiomes and much more in over 25 island schools through the use of a mobile nanopore sequencing lab. A major objective of the program is to advance life sciences and bioethics education in Hawai’i by building capacity and equity across diverse classroom settings, while enabling identity formation and workforce development in STEM careers prior to students entering college. Through the years, we have learned many lessons in response to difficulties around procurement of equipment and reagents, scaling up to larger projects, changes in workflows and chemistries, and limitations in computational tools and expertise. We have also refined our training and onboarding approaches for teachers with varying degrees of prior lab experience through our annual workshops and classroom visits. While some of these challenges and insights are unique to the K-12 setting, others such as the need for classroom-ready bioinformatics solutions beyond the current EPI2ME Labs toolkit are likely more universal across the educational landscape.
Yvonne Chan. As the Director of Community Science at ‘Iolani School in Honolulu, Hawai‘i my mission is to nurture student Independent Research, foster Citizen Science among schools and communities, and provide outreach and workshops to support STEM-education across the state of Hawai‘i. I am trained as a geneticist utilizing ancient DNA technologies for conservation and got my PhD at Stanford University. However, over the last 10 years, I have worked in K-12 education, where my goal has been to energize, mobilize, and engage teachers and students to learn science by doing science in service to their communities. The Office of Community Science has four programs: Our once-a-month teacher learning communities (STEMPlus) have connected >800 teachers, scientists, and community members on a variety of Citizen Science and Technology issues. As one of the project founders and coordinators for Pā‘epā‘e o Waikolu, over the last nine years, we have brought together >25,000 students and >2,000 educators from over 50 educational and professional institutions around the island of O‘ahu to monitor stream biodiversity and water quality and restore our watershed through invasive species removal and stream clean-ups. In 2018 we began the ‘Āina Informatics Network – a network of public, private, and charter schools in Hawai’i bringing genome science and bioethics into high school classrooms. In addition to putting cutting edge technologies into the hands of students, we believe it is critical to prepare students to engage in the bioethical discussions surrounding emerging technologies.
Jessica Chen
Whole Genome Sequencing (WGS) technologies have revolutionized how Public Health and Regulatory Agencies respond to foodborne, waterborne, and one-health related surveillance, detection, and response activities. PulseNet USA, the national molecular surveillance network of over 80 public health labs in the United States, connects food, water, and one-health related illnesses that may be part of an outbreak, fully implemented WGS in 2019. WGS data is comprehensive: antimicrobial resistance, virulence markers, serotype markers, plasmids, and core and accessory genes are all characterized as part of a single WGS workflow. Currently, PulseNet USA sequences approximately 65,000 Salmonella, Escherichia, Listeria, Vibrio, Campylobacter, and Shigella isolates annually. PulseNet International spans 88 countries and seeks to advance molecular surveillance globally by facilitating communication, data sharing, technical assistance, and trainings across the network. Leveraging Oxford Nanopore sequencing in PulseNet International is especially attractive due to ONT’s global accessibility, small laboratory footprint, and cost effectiveness. Through our PulseNet Asia Pacific and PulseNet Africa trainings, we trained participants from 23 countries on ONT sequencing. During this presentation we will discuss our experiences validating this technology for a large surveillance network as well as enabling WGS and providing trainings on ONT and bioinformatics in lower resource settings.
Jessica Chen is the lead of the Bioinformatics Research and Development Team in the Enteric Diseases Laboratory Branch (EDLB), at the Centers for Disease Control and Prevention. Jessica's team provides bioinformatics support for EDLB's current initiatives to validate ONT sequencing for foodborne pathogen surveillance as well as international training activities. Prior to joining CDC, Jessica completed a postdoctoral fellowship in microbial food safety at the University of British Columbia. Jessica received her Ph.D. in Animal Science with an emphasis on microbial food safety from Texas Tech University in 2013. Jessica also holds an M.S. in Animal Science from Colorado State University and a B.S. in Food Science from Cornell University. In her spare time Jessica enjoys spending time with her family, playing board games, hiking Georgia’s parks, and birding.
Lisa Darmo
Carolina Biological Supply has provided educators with materials and support for success in the classroom for nearly one hundred years. We get teachers what they need, when they need it, in the quantities needed. We are proud of our nearly forty-year collaboration with the DNA Learning Center and Dr. Micklos to help bring cutting edge principles and practices of Biotechnology to the classroom. Building on that successful history we are excited to partner with Oxford Nanopore Education and DNALC to help fulfill the visionary promise of Nanopore sequencing in Education. Our Product Development staff is currently working with DNALC and Oxford Nanopore for development of kits tested with teachers. The educational kits are designed for teachers that may have a limited understanding of the use of Nanopore technology. As such they will help fulfill Oxford Nanopore Technologies’ vision to enable analysis of anything, by anyone, anywhere. Through US distribution of Nanopore Education hardware, chemistry and kits we will effectively bring this exciting and unique technology to educators while providing immediate robust and accurate customer service. Our long history of supplying and supporting teachers will help ensure their success with this innovative and accessible technology.
Lisa Darmo. I have been a Department Head and Manager with Carolina Biological Supply since the mid-1990s and have enjoyed being part of the growth and development of our partnership with DNALC. As a scientist and former classroom instructor I am proud of what we have done to bring cutting edge molecular biology concepts to the high school and college classroom. I look forward to participating in this event.
Hayley DeHart
Widespread availability of DNA sequencing platforms has shown great promise for agile research responses, but significant technical implementation challenges continue to limit their deployment. Our group at Johns Hopkins Applied Physics Laboratory is developing capabilities intended for fully autonomous in situ platforms, including sampling hardware, highly optimized molecular biology, microfluidic sample preparation devices, and a bioinformatics software platform for distributed sequencing in low-resource environments. Elements of these capabilities have been deployed for field-based sequencing in locations ranging from Cambodia for highly pathogenic avian influenza to polar oceans for environmental DNA analysis. In this presentation, we will highlight advances that have enhanced our ability to perform in situ sequencing and analysis in resource-constrained environments, including lessons learned as resulting data is used to inform decision-making. Engagements in educational settings across varying levels of skill and experience will also be highlighted as we work towards the goal of developing ubiquitous biosensing capabilities.
Hayley DeHart is a genomics research scientist at the Johns Hopkins Applied Physics Lab (APL). With an educational background in marine biology and a technical background in genomic sequencing methodology, she aims to characterize communities from environmental samples. While at APL, she specializes in marine environmental DNA research and integrating molecular biology approaches with engineering automation capabilities.
Ray A. Enke
Undergraduate students learn about DNA sequencing technology in courses, but often have difficulty understanding the impact of these techniques without hands-on experience. Over the past seven years, undergraduate educators at James Madison University, a 4-year public institution, have developed innovative course-based undergraduate research experiences (CUREs) utilizing Sanger, Illumina, and Nanopore sequencing technologies. An immersive 2 semester CURE leveraging the DNA Barcoding pipeline was implemented into large-enrollment 1st year Foundations of Biology courses. During these experiences, students create DNA barcodes using Sanger sequencing then learn basic bioinformatics skills to analyze their own data. DNA Metabarcoding CUREs stack skills undergraduates learn using the barcoding pipeline in the contexts of high throughput Illumina sequencing applied to 16S microbial metabarcoding experiments. Nanopore long read library prep and sequencing allows students to take part in all steps required to assemble chloroplastic genomes in the context of a genetic incompatibility experiment. Collectively, these CUREs expose undergraduate students to different facets of DNA sequencing technology and applications.
Ray Enke. As an Associate Professor of Biology at James Madison University, my research lab studies transcriptional regulation in retinal neurons. My scholarship also focuses on developing and disseminating novel genomics and proteomics activities for course-based undergraduate research experiences (CUREs). My most current interests involve leveraging Nanopore sequencing for 16S and other amplicon-based DNA metabarcoding course-based research projects.
Linnea Fletcher
The Austin Community College (ACC) Biotechnology Program offers stackable credentials to students starting with an entry level certificate, an Associate of Applied Science degree, and an Advanced Technical Certificate for students who already have a 4-year degree. Students in the high school program complete the entry level certificate. Students in all these pathways complete DNA sequencing projects. Prior to the NSF-ATE Project Award 2055607, students completed upstream protocols for sample prep and samples were set out for sequencing. With the awarding of the ATE grant, DNA sequencing centers for barcoding were established. An immediate benefit was the entire workflow for sequencing projects, from sample to sequence, was owned by the students, making students more interested in working in the industry. Plus, the Industry Biotechnology Advisory Board indicated learning these skills enhanced students’ ability to get jobs in the company. Presently, ACC is seeking additional ATE funding to support the development of Nanopore sequencing based research experiences and industrycertified micro-credentials to further engage our students and verify their competence in this important skill for employment. Students will be able to sequence plasmids used in upstream manufacturing, sequence phage, and expand the genomic based projects. The ACC Biology Department and Environmental Science Department have already purchased Nanopore equipment for their undergraduate research projects.
Dr. Linnea Fletcher enjoys all forms of exercise but especially biking, hiking, and swimming. Her favorite pastimes are family events in the outdoors and travel. She received her Ph.D. in microbiology from the University of Texas at Austin, and did two postdocs: one at the Southwestern Medical Center, and another in the Biochemistry Department at the University of Texas. She joined Austin Community College (ACC) as a Department Chair in Biology and started the Biotechnology Program in 1999. At the same time, she joined the first NSF-funded National Biotechnology Education Center, Bio-Link, and received her first NSF-funded ATE grant to start biotechnology high school programs in Texas. She worked as an NSF Program Officer from 2008 to 2010 and was involved in setting up the first Vision and Change Meeting. In 2015, she received an Emerging Technology Fund Grant to build a bioscience incubator at ACC and several Wagner Peyser Grants to equip it. Today the incubator is full of start-up companies and students interning or working for these companies. She was PI of the AC2 Bio-Link Regional Center, the PI of InnovATEBIO 1.0, and now InnovATEBIO 2.0, the NSFfunded National Biotechnology Center. Combining economic development with educational opportunities is her passion.
Jon Hale
The biology curricula in the UK for 15-18 years olds does not leave space for students to truly explore the world around them. The Daffodil DNA Project provides a mechanism for teachers to bring Nanopore sequencing into the classroom as an anchor for abstract concepts, allowing students opportunities to undertake sequencing whilst learning about cell ultrastructure to phylogeny. In addition to conceptual knowledge, students have changed their attitudes towards STEM subjects and their aspirations as they identify as scientists. Furthermore, as their collaborating teacher identify as scientists, these adults begin modelling the career to all of their students, which may further enhance the science uptake at their respective schools. This project is now working with approximately 30 schools throughout the UK, each with a specific aim to sequence the chloroplast genome of two daffodil cultivars having evolved from a single school’s project in 2020. As this collaboration grows, the limiting factors will be the involvement of partnering scientists willing to provide support and the increasing financial constraints as inflation continues to squeeze budgets.
Jon Hale is a secondary school (11-18 years old) biology teacher at Beaulieu Convent School where he developed the Daffodil DNA Project (2019). In this project students sequence chloroplast DNA using nanopore technology to assess phylogenetic relationships. The project has been successfully scaled up across many schools in the UK. Jon is now analyzing the impact of the project on students, teachers and scientists via a PhD at the University of Dundee alongside his teaching commitments.
Jennifer Katcher
DNA Barcoding enables students to ask and answer their own research questions relevant to their communities - this can include identification of insect vectors like mosquitoes or pathogenic crop pests. Nanopore sequencing allows students to complete all DNA Barcoding steps from start to finish - no more sending away samples for sequencing. Nanopore sequencing could be an extension of two national-scale CUREs - SEA Phages and Tiny Earth - which includes approximately 20,000 students annually. Including Nanopore sequencing in DNA barcoding or metabarcoding curricula will benefit numerous students and provide more equitable research opportunities for students at under-resourced schools. I have designed, implemented and assessed “Bee the CURE '', a successful pollinator DNA barcoding CURE at Pima College, a Hispanic-Serving Community College. I am also a collaborator for the Arecibo C3 project which brings DNA Barcoding to high school and undergraduate students in Puerto Rico. I’ve participated in a DNALC’s “Summer of Nanopore Sequencing” workshop, Oxford Nanopore’s Education Beta Programme and the Nanopore CUREs FMN group through QUBES. I was senior personnel for the “CUREs in HSIs” workshop in 2022 and facilitated sessions in CURE development, assessment and sustainability. I look forward to contributing my experience and enthusiasm for Nanopore sequencing to this group!
Jennifer Katcher has taught undergraduate biology for 20+ years at Pima Community College, a Hispanic-Serving Community College in Tucson, AZ. She champions coursebased research opportunities for students at under-resourced institutions because early access to research is an equity issue. Jennifer leads "Bee the CURE" a communitybased DNA barcoding CURE (Course-based Undergraduate Research Experience) enabling students to identify native bee diversity. Her interest in Nanopore sequencing was immediate since it enables students to complete all steps of DNA barcoding in the classroom. She has participated in the Nanopore Faculty Mentoring Network and the Oxford Nanopore Education Beta Programme. Jennifer is active in CURE faculty mentoring locally and nationally. She serves as the first "STEM CURE Coach" at Pima and contributes to the University of Arizona's CURE Institute. She facilitated sessions in CURE development, assessment and sustainability for the NSF-sponsored “CUREs in HSIs” workshop in 2022. She is also a collaborator for the Arecibo C3 project. Jennifer is a Partnership for Undergraduate Life Science Education (PULSE) Ambassador trained to facilitate biology departments in implementing evidence-based best teaching practices.
Stephen Koury
Our project immerses students and teachers in a citizen science research project focusing on assessing waterway health within the Western New York. It is supported by an NIH Science Education Partnership Award (SEPA) R25 grant. The project promotes the role of genomics education in workforce development by informing students, educators and other relevant organizations in the ways genomics integrates into future STEM jobs and careers. Participants use Oxford Nanopore MinION sequencing technology to get first-hand experience with the preparation of genomic DNA samples for sequencing, in school DNA sequencing, tools for determining the makeup of microbial communities from sequencing data, and the construction of a microbial genome through the compilation of overlapping long MinION sequence reads. The project includes a weeklong teacher professional development covering hands on experience with all aspects of project activities in the summer. Students then perform identical project activities during the academic year under the guidance of their teachers and partnership faculty. Students and teachers then present their data at an annual capstone event each spring. A summary of project activities through the first 4 years of the award will be presented.
Stephen Koury. I have been an educator/researcher in the Department of Biotechnical and Clinical Laboratory Sciences in the Jacobs School for over 30 years. My B.S. in Medical Technology and M.S. in Biology were done at the University at Albany and my Ph.D. was done in the department of Anatomical Sciences at the University at Buffalo. Post-doctoral training was done at Vanderbilt University in the Hematology Division of the Department of Medicine. Research interests have generally been in the areas of erythropoiesis, general molecular biology, and, most recently, microbial genomics. The department of BCLS houses B.S. programs in Medical Laboratory Sciences and Biotechnology, as well as an M.S. program in Biotechnology. Courses taught include Biomolecular Technology and Diagnostics, Introduction to Human Medical Genetics, Introduction to Microbial Genome Annotation and Applications of Molecular Biotechnology. Of relevance to this meeting are past NIH SEPA and NSF ITEST grants as a Co-PI involving basic bioinformatics, as well as the current SEPA dealing with metagenomic and whole genome sequencing. I am very enthusiastic about bringing new STEM technology experiences to students in hopes of igniting an interest in science that many of my teachers in my grades 6-12 did for me.
Sebastian Kraves
The Genes in Space science competition challenges U.S. students in grades 7-12 to design molecular experiments addressing the biological challenges of living in space. Through a freely accessible online portal students submit proposals that undergo rigorous selection, active mentorship and, for one annual winner, implementation by astronauts aboard the International Space Station (ISS). Four student winners proposed to investigate how DNA damage puts spacefarers at increased risk of negative health consequences including cancer. Using CRISPR to induce targeted DNA lesions that mimic genetic damage caused by cosmic radiation, students would identify the DNA repair pathways activated in space. Their proposal was carried out aboard the ISS by astronauts Christina Koch and Nick Hague in 2019. Following DNA lesions in eukaryotic cells, Nanopore sequencing revealed the DNA repair pathway choices made in space. Understanding how innate DNA repair mechanisms respond to space conditions is a step toward designing safeguards for future space travelers. The technology’s accessibility enabled this study to carry out a complex molecular biology workflow in microgravity: In addition to Nanopore sequencing, it involved genetically transforming and culturing cells, amplifying microbial DNA via PCR, and performing the first CRISPR gene editing experiment in space.
Dr. Sebastian Kraves co-founded the Cambridge-based start-up miniPCR bio to help bring DNA analysis technology to the masses. Kraves was previously a principal at BCG, where he spent more than six years working on health care challenges, such as how to make biomedical technology accessible in sub-Saharan Africa. A molecular neurobiologist who trained at Harvard, Kraves is a patented inventor who has published research on optogenetics and the genetic regulation of behavior, but is now focused on his dream to make DNA analysis tools accessible to everyone, everywhere. In 2015, Dr. Kraves co-founded Genes in Space, a collaboration between miniPCR bio and Boeing with support from ISS National Lab and New England Biolabs. This initiative invites youth to design innovative space biology experiments to be conducted aboard the International Space Station (ISS). Genes in Space has flown ten breakthrough student-led research investigations to the ISS, eight of which have resulted in peer-reviewed publications led by high school students. Dr. Kraves is committed to keep fueling their dreams.
Rebecca Lewis
The Amgen Biotech Experience (ABE) is an innovative outreach program designed to introduce secondary school students to molecular biology and biotechnology through hands-on laboratory experiences. ABE provides educational materials, training and support for teachers, and state-of-the-art laboratory equipment loans to schools, with the goal of enhancing students' understanding of complex biological concepts and the practical applications of biotechnology. Outreach programs like ABE are crucial for expanding access to advanced scientific education, particularly in underserved communities. ABE democratizes learning by bringing cutting-edge technologies into classrooms that might otherwise lack such resources. Additionally, these programs can increase awareness and knowledge of sequencing technologies, which are pivotal in modern biological research and medicine. By integrating these technologies into secondary school curricula, ABE and similar initiatives not only foster a deeper understanding of molecular biology but also inspire future generations of scientists and innovators. Such programs bridge the gap between high school education and higher-level scientific careers, contributing to a more informed and skilled workforce in biotechnology and related fields.
Rebecca Lewis leads initiatives that catalyze young people’s interest in STEM and connect classroom experiences with the world of work. She brings extensive experience in instructional design, interdisciplinary learning and teaching, professional development, science education, enhancing formal and informal STEM learning, broadening participation in STEM, and effective strategies to promote college and career success.
An experienced senior manager, Lewis leads initiatives that focus on engaging leaders from STEM industries, corporations, schools, and communities in collaborating to improve students’ outcomes. She manages a large portfolio of projects focused on strengthening STEM education in the U.S. and abroad. As director of EDC’s Amgen Biotech Experience (ABE) Program Office, she provides opportunities for teachers to bring biotechnology to their classrooms at no cost. ABE provides access to cutting-edge science through four key areas: high-quality teaching and learning resources; professional learning experiences, access to a global community of practice, and implementation resources for teachers.; connecting scientists to educators and classrooms; and providing opportunities for underserved and underrepresented populations.
Alice Matimba
Wellcome Connecting Science’s Learning and Training programme is a global leader in advancing genomics, engaging 20,000+ scientists annually. Offering a curriculum covering human, pathogen and other organisms, it delivers 60+ events each year, comprising in-person courses, conferences, MOOCs, and blended learning formats. Through these activities, the programme plays a critical role in workforce development and building sustainable genomic research capacity globally. A key component of the programme is providing training in regions such as Africa, Asia, and Latin America. By integrating cutting-edge technologies like Nanopore within courses, trainees are provided with hands-on experience, enabling them to apply real-time, portable sequencing solutions in their work. This training democratises access to genomics, promoting equity by ensuring under-resourced settings can participate in this rapidly evolving field. By incorporating accessible sequencing technologies and data analysis approaches within courses, the programme serves as a catalyst for expanding the practical application of genomics on a global scale. However, the success of these initiatives isn’t solely dependent on effective education. To achieve longer-term impact, trainees must apply their learning in practice, necessitating access to sequencing resources post-training. Strategic collaborative partnerships with commercial companies are crucial in overcoming these challenges, ensuring genomics training translates into real-world outcomes.
Dr. Alice Matimba is the Head of Training and Global Capacity at Wellcome Connecting Science, where she leads initiatives to enhance genomics skills and build capacity worldwide. She holds a PhD in Human Genetics from the University of Cape Town, South Africa and completed postdoctoral research at the Mayo Clinic, USA. Dr. Matimba has a distinguished career in biomedical science, having supervised postgraduate students, led research projects, and authored over 40 peer-reviewed publications. Her research focuses on the genomics of African populations, functional pharmacogenomics, public health interventions for non-communicable diseases, and ethical issues. In addition to her scientific contributions, she is the Executive Producer of the Your Digital Mentor Podcast, a series that explores mentoring and career development through real stories and expert insights.
Nirav Merchant
AI-powered platforms are reducing obstacles to data exploration with innovative analysis methods. They free users from the need to master complex syntax or programming languages before using these powerful methods. AI is revolutionizing how we design, build, utilize and teach with these capabilities and platforms. However, the real cost and accessibility of many of the underlying proprietary technologies (GPT, LLM etc.) are rapidly widening the digital divide, where you need a subscription to perform meaningful tasks. We all have the collective responsibility to ensure that these tools and platforms are developed with equity, efficiency, and affordability in mind through their lifecycle. I will highlight efforts undertaken by CyVerse and other NSF projects, federal agencies, and commercial organizations for ensuring access to sustainable AI-based infrastructure.
Nirav Merchant serves as the Director of the Data Science Institute at the University of Arizona; his research has been focused on development of scalable computational platforms for open science.
Sarah Miller
Tiny Earth addresses two pressing challenges: antibiotic resistance and a diverse STEM workforce. Tiny Earth is a global CURE with 800+ trained instructors in 33 countries. The goal of the network is to discover new antibiotics from soil bacteria and encourage diverse students to persist in STEM. The flexible curriculum costs no more to implement than a traditional microbiology course lab and focuses on hypothesis-driven research skills. The companion instructor training (a week-long immersive, facilitated institute at UW-Madison) is based on scientific teaching practices and AJEDI principles. This presentation will provide an overview of the Tiny Earth curriculum and network. It will also show how we address our goals as a global network, secure public and private contributions, and keep equity and inclusion at the center of everything we do.
Sarah Miller is a National Academies Education Mentor in the Life Sciences and received the UW-Madison Teaching Academy Distinguished Teaching Award. She is the Executive Director of Tiny Earth at UW-Madison, overseeing an international network of 800+ instructors in 33 countries who teach an estimated 16,000 students annually. Tiny Earth is a coursebased undergraduate research experience (CURE). The goal of Tiny Earth is to studentsource the discovery of new antibiotics from soil bacteria and increase diversity in STEM. Sarah’s professional interests focus on STEM education, with an emphasis on active and inclusive learning, institutional transformation at scale, and faculty/graduate development in higher education. She co-authored five publications in Science magazine, in addition to other education journals, most recently two articles in JMBE about leveraging communities of practice to create antiracist and online curriculum for research courses. She founded the Scientific Teaching Book Series and is a co-author of Scientific Teaching and Entering Mentoring. During her graduate work in plant pathology at UW-Madison, she investigated the environmental impact of genetically modified organisms (GMOs) by analyzing microbial communities affiliated with plant rhizospheres.
Jonathan Pugh
Oxford Nanopore Education aims to make a profound impact on society by embracing inclusivity and equipping the next generation of scientists. We approach this through two methods: 1) enabling our Community and helping to amplify their efforts in the classroom, and 2) partnering with education institutions to combine their expertise with our unique tools. In this talk I will cover our progress on these fronts, and our future plans to deliver accessible, equitable science.
Jonathan Pugh has devoted over a decade to Oxford Nanopore Technologies, advancing from a foundational role in nanopore research to currently spearheading the company's educational initiatives as the Director of Nanopore Education. With a rich career encompassing product management, marketing, and now education, Jonathan leverages his extensive expertise to bring sequencing technology to a wider student demographic across the globe. His current role reflects a harmonious blend of his adept communication skills and a deep-seated knowledge of nanopore sequencing, aiming to foster STEM engagement and education for the next generation of scientists.
Pushpa Ramakrishna
The SEA-PHAGES program provides a two-semester sequence of discovery-based authentic research experiences for students to gain hands-on experience in microbiology, molecular biology, and bioinformatics. It engages thousands of undergraduate students in the discovery and characterization of bacteriophages. This program provides faculty and students an opportunity to collaborate and contribute to a growing repository of phage genomes that are valuable for understanding microbial diversity and evolution. Participation in the SEAPHAGES inclusive Research Education Community (iREC) has found profound impacts on students' sense of belonging, persistence, success, and agency in science.
Dr. Pushpa Ramakrishna is a program officer at the Center for the Advancement of Science Leadership and Culture at the Howard Hughes Medical Center (HHMI). She was previously at the NSF’s Division of Undergraduate Education as a program officer. She is also an emeritus professor at Maricopa Community Colleges where she led many initiatives on STEAM and Sustainability education, undergraduate research, service learning and other high impact practices. She is passionate about bringing in systemic change in undergraduate STEM education through the lens of equity and inclusivity. She strives to bring the excitement from the frontiers of research into the undergraduate classroom for all students.
Laura K. Reed
Actual research experience as an undergraduate is one of the best ways to support retention and meaningful learning in STEM, however there are many barriers for most undergraduates at US institutions to having those experiences. In addition to various financial and logistical barriers, there are barriers in faculty expertise and confidence in teaching about cutting-edge genomics methodologies. Most faculty received their graduate training before the current technologies and analysis methods were developed. The Genomics Education Partnership (thegep.org) is one example of a successful effort to bring genomics research to thousands of undergraduates through course-based undergraduate research experiences (CUREs). The GEP’s success is largely due to its community of practice that give faculty confidence to lead students in genomics research. Through the GEP, faculty develop and maintain a current understanding of the field, are supported through the challenges of running a CURE, are provided up-to-date curriculum, and gain professional advancement through research publication. Historically, the GEP has focused on in-silico research, however the partnership is now exploring cost effective genomic data generation in the classroom. The lessons the GEP has learned about supporting faculty will be valuable as the field brings de novo genomics data generation to the undergraduate classroom.
Laura K. Reed is a Professor of Biology at the University of Alabama. Dr. Reed received her BS in Biology (University of Oregon), her PhD in Ecology and Evolution (University of Arizona), was NIH-NRSA postdoctoral fellow (North Carolina State University), and joined the faculty at the University of Alabama in 2010. Her research focuses on the evolution of complex traits and effective science education methodology. She joined the GEP in 2011, taking on the role of Director in 2017. Under Dr. Reed, the GEP has more than doubled its faculty membership, enhanced its educational resources, and broadened its scientific scope.
Robert Rivers
The presentation will provide a brief overview of current activities and work at the National Human Genome Research Institute aimed at developing a robust genomics workforce reflective of the nation. The presentation also poses questions for consideration of how novel technology combined with considered investment can be applied to further democratize opportunities.
Robert Rivers is currently the director of the Training, Diversity and Health Equity Office (TiDHE) at the National Human Genome Research Institute. The TiDHE Office works to champion a diverse genomics workforce, address health disparities and reach a more equitable and healthy society. He previously served as the Acting Director of the Office of Minority Health Research Coordination at the National Institute of Diabetes and Digestive and Kidney Disease. There he assisted program staff and officials in advancing programs and initiatives that address diseases and disorders that disproportionately impact the health of racial and ethnic minority populations and promote programs that fosters the recruitment and training of individuals from diverse backgrounds. Previously he served as a Program Official at the National Cancer Institute and joined the federal government through the AAAS Science & Technology Policy Fellows program. Robert received a B.S. in Chemistry from Kentucky State University, and a Ph.D. in Chemistry from University of Cambridge.
Dan Russell
Each year, SEA-PHAGES students around the country discover thousands of new phages and send hundreds of them to be sequenced at the University of Pittsburgh. Because the program has been running for 15 years, these phages have been sequenced by several different technologies, including Sanger, 454, IonTorrent, PacBio, Illumina, and Oxford Nanopore. This sequencing history has provided us with unique experience to assess the benefits and drawbacks of a variety of data types as they pertain, in particular, to small genomes. We will discuss how Oxford Nanopore sequencing is currently used in the SEA-PHAGES program, and how its low entry cost and ease of use might expand opportunities for SEA-PHAGES students.
Dan Russell graduated from Cornell University with a degree in Microbiology, then went to work at The Institute for Genomic Research where he learned both the wet bench and bioinformatics sides of sequencing and genomics. He later earned a Master's in Education from the University of Missouri, taught high school science for several years, then moved to Graham Hatfull's group at the University of Pittsburgh where he's been for the last 15 years. Dan has experience with a variety of sequencing technologies and data, and has probably sequenced more individual bacteriophage genomes than anyone else in the world.
Susanna Theroux
Molecular (DNA- and RNA-based), methods have revolutionized our ability to monitor and assess biological communities with greater speed, accuracy, and precision. However, many regional monitoring programs are struggling to incorporate a molecular component into their routine monitoring due to difficulties in translating DNA-based taxonomy to microscopy-based scoring tools as well as limited access to sampling and analytical equipment and expertise. We will review efforts to expediate the incorporation of molecular methods into targeted species monitoring and community bioassessment programs in California and the potential for nanopore technology to further advance molecular method adoption.
Susanna Theroux, PhD is a principal scientist at the Southern California Coastal Water Research Project (SCCWRP) where she works on the development of eDNA methods for biomonitoring and bioassessment. She also leads the California Molecular Methods Workgroup and the West Coast Ocean Biomolecular Observing Network.
Spike Willcocks
Oxford Nanopore Technologies commercialized the world’s first nanopore-based sequencing device in 2015 after decades of research from industrial and academic communities. Jumping to present day this technology has been deployed across the globe for a multitude of applications, delivering insights previously not possible through any other sequencing technology. In this talk Spike will cover the origins of Oxford Nanopore, the journey from idea to product, and how the businesses and partnerships now utilizing this technology will impact the development of our future workforce.
Spike Willcocks. After studying Chemistry at the University of Oxford and completing a D.Phil. in Biochemistry, Spike started working for the intellectual property investment business “IP Group” in 2001 as an early employee. He ultimately led its Life Sciences group, and was responsible for the formation of Oxford Nanopore Technologies during that time. Gordon Sanghera persuaded Spike to join Oxford Nanopore full-time at the start of 2006, leading the Corporate and Business development function, and now he is the company’s Chief Strategy Officer.
Jason Williams
Genomes aren’t just at the center of cells; they are at the center of ethical, social, and policy issues that determine who benefits from genomics and who is excluded. As truly accessible genomics technology enters the classroom, a holistic approach to pedagogy and research practice can showcase why genomics research experiences are a uniquely powerful vehicle for rigorous, impactful, and inclusive STEM education. Equity issues in genomics are well-documented. At the geopolitical level, the genomes that have been sequenced overrepresent the interests of Western populations and affluent regions. Health policy and intellectual property rights are shaped by this bias, which in turn leads to disparities such as those seen in clinical trial participation and ownership of agricultural biotechnology. Correspondingly, research and education for privileged groups provide their members with the equipment and training not just to learn what genomics is, but to work with genomes. This inequitable cycle continues as privileged students enter the workforce with the ability to shape genomics to their benefit. Thinking holistically about genomics education could subvert this cycle in favor of a more equitable and inclusive outcome. For STEM students, holistic genomics education should adhere to two minimum requirements: first, that genomics education includes both molecular and data science techniques through meaningful research experiences; and second, that it incorporates responsible conduct of research practices and culturally relevant pedagogy that contextualize the practice and implications of genomics work. Achieving this standard would entail efforts on two fronts: first, establishing a national-scale support system to provide accessible, low-cost genome research experiences—including equipment, curricula, and professional development; and second, ensuring that curricular and professional development, as well as research outputs, support culturally relevant pedagogy and promote responsible research principles that foster ethical and nonexploitative learning outcomes and research products.
Mr. Jason Williams is Assistant Director, Inclusion and Research Readiness at the Cold Spring Harbor Laboratory DNA Learning Center where he develops national biology education programs. Mr. Williams has delivered professional development and training for thousands of students, researchers and educators in bioinformatics, data science, and molecular biology. His focus has been developing bioinformatics in undergraduate education and career-spanning learning for biologists. Jason is founder of LifeSciTrainers.org – a global effort to promote community of practice among professionals who develop short-format training for life scientists. Jason is advisory to cyberinfrastructure, bioinformatics, and education projects and initiatives in the US, UK, Europe, and Australia.
Anna Feitzinger, Dave Micklos, Jeffry Petracca, Jason Williams
Cold Spring Harbor Laboratory DNA Learning Center
DNA Subway is one of the most widely used education bioinformatics platforms for analyzing DNA sequence— with 37,000 users logging 84,000 sessions in 2023. Over 270,000 cumulative sequences, primarily DNA barcodes, have been uploaded from our sequencing partner GENEWIZ. Now, the growing popularity of nanopore sequencing adds a new data source for DNA Subway. The Blue Line is optimized for analyzing DNA barcodes and supports course-based undergraduate research experiences (CUREs), independent student research, and citizen science projects on biodiversity. The Blue Line has tools for uploading and editing barcode sequences, developing consensus sequences, performing BLAST searches and multiple sequence alignments, and generating phylogenetic trees. Through a grant from the NSF Advanced Technological Education Program, DNA Subway 2.0 is being redeveloped as a mobile-first and accessible application. We believe this will be the first bioinformatics workflow optimized for use on cell phones. We have added new tools for importing Nanopore sequence data to the Blue Line workflow. In combination with an Oxford Nanopore device, DNA Subway 2.0 will deliver on the promise of sequencing and analyzing DNA barcodes in the field, in school, and at home—by anyone, at any time.
Anna Feitzinger, Ph.D. is Assistant Director for Science of the DNA Learning Center of Cold Spring Harbor Laboratory, a science center devoted entirely to public genetics education. She applies her expertise in quantitative biology and biochemistry in bridging the gap in bioinformatics education at the pre-college and college level. She is devoted to making DNA sequencing accessible in education, with the dream that Oxford Nanopore sequencers will be available to students in classrooms worldwide. She received a B.A. in Chemistry from the City University of New York, Hunter College, and a Ph.D. in Biology at the University of California, Davis.
Dave Micklos is the Executive Director of the DNA Learning Center (DNALC) at Cold Spring Harbor Laboratory. Dave founded the DNALC in 1988 as the world’s first science center devoted to public genetics education. The DNALC has since grown to five sites in the New York Metropolitan area, encompassing 45,000 square feet, 16 teaching labs, and four bioinformatics labs. Nineteen additional programs at universities and scientific institutions worldwide have been licensed by or modeled after the DNALC. Dave’s career has focused on developing large-scale lab and computer infrastructure to allow large numbers of students and citizen scientists to inquire about the genetic basis of life. The DNALC first implemented footlocker kits and mobile labs to costeffectively carry DNA experiments around the world and developed the first student experiment in personal DNA sequencing. The DNALC’s methods for DNA barcoding and its DNA Subway bioinformatics platform form one of the most widely used systems for undergraduate student research and citizen science studies of biodiversity. More than 500,000 US students per year use curricula, experiments, and extended research projects developed by the DNALC.
Jeffry Petracca. I am the Manager of Student and Public Research at the DNA Learning Center. I've worked at the DNALC for more than seven years where I teach many of the high school camps and courses. Before the DNALC, I worked for more than 20 years in the zoological field where I most recently served as the Curator of Entomology at the Long Island Aquarium in Riverhead, NY. As a trained entomologist, I'm also responsible for curating many of the DNA barcodes generated via the DNALC's citizen science initiatives for publication to GenBank. Along with my colleagues at the DNALC, I've coordinated and provided guidance for a number of student and citizen scientist experiments that utilize ONT throughout the United States and at our partner DNA Learning Center in Suzhou, China. My experience with ONT focuses on its application for DNA barcoding, metabarcoding, and DNA sequencing in the classroom. I am very interested in bringing this technology to the field and to nature reserves around the world seeking to involve their local community in learning about the union of conservation and biotechnology.
Mr. Jason Williams is Assistant Director, Inclusion and Research Readiness at the Cold Spring Harbor Laboratory DNA Learning Center where he develops national biology education programs. Mr. Williams has delivered professional development and training for thousands of students, researchers and educators in bioinformatics, data science, and molecular biology. His focus has been developing bioinformatics in undergraduate education and career-spanning learning for biologists. Jason is founder of LifeSciTrainers.org – a global effort to promote community of practice among professionals who develop short-format training for life scientists. Jason is advisory to cyberinfrastructure, bioinformatics, and education projects and initiatives in the US, UK, Europe, and Australia.
This meeting is supported by grants from the National Science Foundation: Improving Undergraduate STEM Education (#1821657), and Advanced Technological Education (#1901984). Additional travel support is provided by Oxford Nanopore Technologies.


